PUmPER: Phylogenies Updated PERpertually
About PUmPER
Fernando, Stephen Smith, and John Cazes have developed PUmPER, a Ruby framework for phylogenetic analyses. It can be used to build a pipeline that automatically updates reference trees when enough new sequences for the clade of interest appear on GenBank.
The tool uses RAxML-Light to extend trees and Stephen's PHLAWD pipeline to extend alignments by new sequences. The code can be run on stand-alone servers and on cluster systems.
Availability
The prototype version including documentation is available via Fernando's github repository.Workflow for an iteration
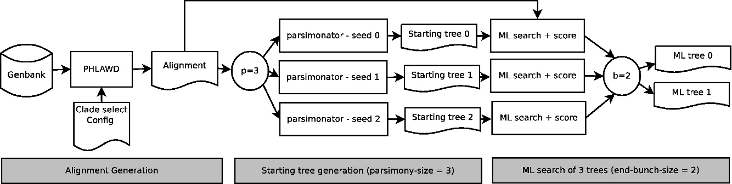
An initial iteration of the pipeline:
- PUmPER constructs an alignment comprising sequences from a clade (from the NCBI taxonomy) and gene regions of interest specified by the user.
- The alignment is then used to infer Maximum Likelihood trees in parallel.
- The best trees are collected and kept to be re-used as starting trees on the next iteration.